Split plot and strip plot analysis in R studio
What is Split Plot design?
A split plot design is a special case of a factorial
treatment structure. It is used when some factors are harder (or more
expensive) to vary than others. In simple terms, a split-plot experiment is a
blocked experiment, where the blocks themselves
serve as experimental units for a subset of the factors.Thus, there are two levels of experimental units. The
blocks are referred to as whole plots, while the experimental units within blocks are called split plots, split
units, or subplots. Corresponding to the two levels of
experimental units are two levels of randomization. One randomization is conducted to determine the
assignment of block-level treatments to whole plots.
Then, as always in a blocked experiment, a randomization of treatments to split-plot experimental units
occurs within each block or whole plot.
Split plot design has a mixture of hard to randomize ( hard to change) and easy to randomize (easy to change) factors. The hard to change factors are implemented first and followed by easy to change factors. It was invented by Fischer in 1975. This type of design is seen in agriculture, industries and medical fields. It is appropriate for all those studies where you find it difficult to change or randomize a factor. Split plot design is generally used when factors are not of same importance.
Example. An experiment is to compare the yield of three
varieties of Moong bean (factor A with a=3 levels) and four different
levels of manure (factor B with b=4 levels). Suppose 6 farmers
agree to participate in the experiment and each will designate a
farm field for the experiment (blocking factor with s=6 levels).
Since it is easier to plant a variety of moong bean in a large field, the
experimenter uses a split-plot design as follows:
- To divide each block into three equal sized plots (whole plots), and each plot is assigned a variety of oat according to a randomized block design.
- Each whole plot is divided into 4 plots (split-plots) and the four levels of manure are randomly assigned to the 4 splitplots.
- We call varieties level the whole-plot factor and manure level as the split-plot factor.
- Whole plots are the experimental units for whole plot factor and split plots for split plot factor.
Note: Observation in the same whole plot shares whole plot error. Precision is more in this type.
For every “level” (whole-plot / split-plot) of the experiment
we have to introduce a corresponding random effect
(better terminology: error) which acts as the experimental
error on that level. To identify the correct design we have to know the
randomization procedure. The general situation can be very complex, but by
following the different randomization levels/steps, setting
up a model is easy.
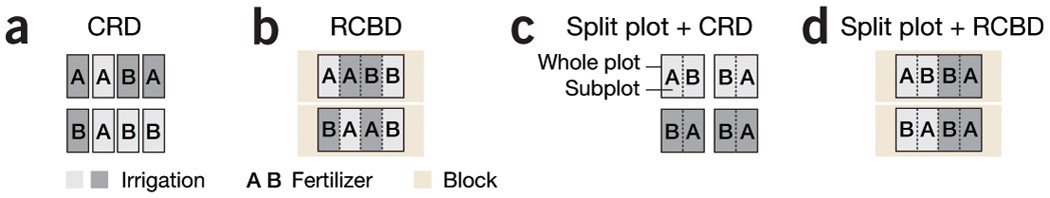
a) In CRD, levels of irrigation and fertilizer are assigned to plots of land (experimental units) in a random and balanced fashion. (b) In RCBD, similar experimental units are grouped (for example, by field) into blocks and treatments are distributed in a CRD fashion within the block. (c) If irrigation is more difficult to vary on a small scale and fields are large enough to be split, a split plot design becomes appropriate. Irrigation levels are assigned to whole plots by CRD and fertilizer is assigned to subplots using RCBD (irrigation is the block). (d) If the fields are large enough, they can be used as blocks for two levels of irrigation. Each field is composed of two whole plots, each composed of two subplots. Irrigation is assigned to whole plots using RCBD (blocked by field) and fertilizer assigned to subplots using RCBD (blocked by irrigation).
Source: https://www.nature.com/articles/nmeth.3293/figures/1
Split plot designs are usually used with factorial sets when the assignment of treatments at random can cause difficulties
– large scale machinery required for one factor but not another
• irrigation
• tillage
– plots that receive the same treatment must be grouped together
• for a treatment such as planting date, it may be necessary to group treatments to facilitate field operations
The split plot is a design which allows the levels of one factor to be applied to large plots while the levels of another factor are applied to small plots
– Large plots are whole plots or main plots
– Smaller plots are split plots or subplots
Levels of the whole-plot factor are randomly assigned to the main plots, using a different randomization for each block (for an RBD). Levels of the subplots are randomly assigned within each main plot using a separate randomization for each main plot.
ü Because
there are two sizes of plots, there are two experimental errors - one for each
size plot
ü Usually
the sub plot error is smaller and has more df
ü Therefore
the main plot factor is estimated with less precision than the subplot and
interaction effects
ü Precision
is an important consideration in deciding which factor to assign to the main
plot
Advantages
ü Permits
the efficient use of some factors that require different sizes of plot for
their application
ü Permits
the introduction of new treatments into an experiment that is already in
progress
Disadvantages
ü Main
plot factor is estimated with less precision so larger differences are required
for significance – may be difficult to obtain adequate df for the main plot
error
ü Statistical
analysis is more complex because different standard errors are required for
different comparisons
Design of Split plots:
There are two blocks, 3 main plot factors (varieties) and 4 subplot factors (varieties) in the figure above.
ANOVA for split plots
Strip plots are also called as split-block design. For experiments involving factors that are difficult to apply to small plots. There are three sizes of plots so there are three experimental errors. The interaction is measured with greater precision than the main effects. For example:
F ratios in Split plot design
ü F
ratios are computed somewhat differently because there are two errors
ü FR=MSR/MSEA tests the effectiveness of blocking
ü FA=MSA/MSEA tests the sig. of the A main effect
ü FB=MSB/MSEB tests the sig. of the B main effect
ü FAB=MSAB/MSEB
tests the sig. of the AB interaction
Variations of split plot design
ü Split-plot
arrangement of treatments could be used in a CRD or Latin Square, as well as in
an RBD
ü Could
extend the same principles to accommodate another factor in a split-split plot
(3-way factorial)
ü Could
add another factor without an additional split (3-way factorial, split-plot
arrangement of treatments)
What is Strip plot design?
Strip plots are also called as split-block design. For experiments involving factors that are difficult to apply to small plots. There are three sizes of plots so there are three experimental errors. The interaction is measured with greater precision than the main effects. For example:
Ø Three
seed-bed preparation methods
Ø Four
nitrogen levels
Ø Both
factors will be applied with large scale machinery
Advantages
ü
Permits efficient application of factors that
would be difficult to apply to small plots
Disadvantages
ü
Differential precision in the estimation of
interaction and the main effects
ü Complicated statistical analysis
ANOVA for strip plot
In
strip plot design, the desired precision for measuring the interaction effect
between the two factors is higher than that for measuring the main effect of
either one of the two factors. This is accomplished with the use of three plot
sizes:
ü Vertical strip plot
for the first factor also called the vertical factor.
ü Horizontal strip plot
for the second factor also called the horizontal factor.
ü Intersection plot for
the interaction between the two factors.
The vertical strip and
horizontal strip plot are always perpendicular to each other. However, there is
no relationship between their sizes, unlike the case of main plot and subplot
of the split-plot design. In this case, the interaction plot is, of course, the
smallest.
Split plot vs strip plot
3
varieties and 4 fertilizer dose are considered for experiment. The varieties
are taken as main plots whereas fertilizers as subplots. Then the layout for
one block indicates the difference between split plot and strip plot as shown
below:
File used: split.xls https://drive.google.com/file/d/1eGIeKlXWHpnPqeTiihsVcGIQAznM6jZu/view?usp=sharing
Note: The output are shown in blue color.
1. Import and attach data set
> split <- read_excel("D:/google/data analysis/r/split.xls")
> attach(split)
2. Observe your data
> head(split)
# A tibble: 6 x 7
Rep Fa Fb Ph1 pdi ch1 ch2
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 R1 Ganesh Jholmal 6.1 0.777 51.5 31.5
2 R1 Ganesh Azadirachtin 5.38 0.999 40.6 36.4
3 R1 Ganesh Imidacloprid 6.28 0.624 43.0 39.0
4 R1 Ganesh Cow's milk 3.55 0.777 46.2 38.1
5 R1 Ganesh Control 3.68 0.777 45.9 35.1
6 R1 Bramha Jholmal 3.9 0.555 46.2 40.7
Note: The factor A and B are set as characters
3. Set Characters as factors
> split$Fa <- as.factor(split$Fa)
> split$Fb <- as.factor(split$Fb)
> str(split)
tibble [75 x 7] (S3: tbl_df/tbl/data.frame)
$ Rep: chr [1:75] "R1" "R1" "R1" "R1" ...
$ Fa : Factor w/ 5 levels "Bishnu","Bramha",..: 3 3 3 3 3 2 2 2 2 2 ...
$ Fb : Factor w/ 5 levels "Azadirachtin",..: 5 1 4 3 2 5 1 2 4 3 ...
$ Ph1: num [1:75] 6.1 5.38 6.28 3.55 3.67 ...
$ pdi: num [1:75] 0.777 0.999 0.624 0.777 0.777 ...
$ ch1: num [1:75] 51.5 40.6 43 46.2 45.9 ...
$ ch2: num [1:75] 31.5 36.4 39 38.1 35.1 ...
Note: Factor A and B have been converted into factors.
4. Now run command for ANOVA
> library(agricolae)
> aov<-with(split,sp.plot(Rep,Fa,Fb,ch2))
ANALYSIS SPLIT PLOT: ch2
Class level information
Fa : Ganesh Bramha Riddi Bishnu Laxmi
Fb : Jholmal Azadirachtin Imidacloprid Cow's milk Control
Rep : R1 R2 R3
Number of observations: 75
Analysis of Variance Table
Response: ch2
Df Sum Sq Mean Sq F value Pr(>F)
Rep 2 163.45 81.73 1.8723 0.2152758
Fa 4 1816.36 454.09 10.4031 0.0029443 **
Ea 8 349.20 43.65
Fb 4 737.16 1 84.29 6.1032 0.0006241 ***
Fa:Fb 16 561.12 35.07 1.1614 0.3379647
Eb 40 1207.83 30.20
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
cv(a) = 16 %, cv(b) = 13.3 %, Mean = 41.40167
Interpretation: The chlorophyll content is significantly different (P<0.001) for both factors i.e. main factor and split factor.
Note: Though the significant difference was not seen for interaction effect, I will be showing the analysis and output for you.
Interpretation: See grand mean, CV, LSD value and which variety is significantly different? ( I have made those values bold).
Note: The output are shown in blue color.
1. Import and attach data set
> split <- read_excel("D:/google/data analysis/r/split.xls")
> attach(split)
2. Observe your data
> head(split)
# A tibble: 6 x 7
Rep Fa Fb Ph1 pdi ch1 ch2
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 R1 Ganesh Jholmal 6.1 0.777 51.5 31.5
2 R1 Ganesh Azadirachtin 5.38 0.999 40.6 36.4
3 R1 Ganesh Imidacloprid 6.28 0.624 43.0 39.0
4 R1 Ganesh Cow's milk 3.55 0.777 46.2 38.1
5 R1 Ganesh Control 3.68 0.777 45.9 35.1
6 R1 Bramha Jholmal 3.9 0.555 46.2 40.7
Note: The factor A and B are set as characters
3. Set Characters as factors
> split$Fa <- as.factor(split$Fa)
> split$Fb <- as.factor(split$Fb)
> str(split)
tibble [75 x 7] (S3: tbl_df/tbl/data.frame)
$ Rep: chr [1:75] "R1" "R1" "R1" "R1" ...
$ Fa : Factor w/ 5 levels "Bishnu","Bramha",..: 3 3 3 3 3 2 2 2 2 2 ...
$ Fb : Factor w/ 5 levels "Azadirachtin",..: 5 1 4 3 2 5 1 2 4 3 ...
$ Ph1: num [1:75] 6.1 5.38 6.28 3.55 3.67 ...
$ pdi: num [1:75] 0.777 0.999 0.624 0.777 0.777 ...
$ ch1: num [1:75] 51.5 40.6 43 46.2 45.9 ...
$ ch2: num [1:75] 31.5 36.4 39 38.1 35.1 ...
Note: Factor A and B have been converted into factors.
4. Now run command for ANOVA
> library(agricolae)
> aov<-with(split,sp.plot(Rep,Fa,Fb,ch2))
ANALYSIS SPLIT PLOT: ch2
Class level information
Fa : Ganesh Bramha Riddi Bishnu Laxmi
Fb : Jholmal Azadirachtin Imidacloprid Cow's milk Control
Rep : R1 R2 R3
Number of observations: 75
Analysis of Variance Table
Response: ch2
Df Sum Sq Mean Sq F value Pr(>F)
Rep 2 163.45 81.73 1.8723 0.2152758
Fa 4 1816.36 454.09 10.4031 0.0029443 **
Ea 8 349.20 43.65
Fb 4 737.16 1 84.29 6.1032 0.0006241 ***
Fa:Fb 16 561.12 35.07 1.1614 0.3379647
Eb 40 1207.83 30.20
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
cv(a) = 16 %, cv(b) = 13.3 %, Mean = 41.40167
Interpretation: The chlorophyll content is significantly different (P<0.001) for both factors i.e. main factor and split factor.
Note: Though the significant difference was not seen for interaction effect, I will be showing the analysis and output for you.
5. Now perform mean separation
a. For Main effect (variety)
> out<-with(split,duncan.test(ch2,Fa,DFerror = 8,MSerror = 43.65))
> out
$statistics
MSerror Df Mean CV
43.65 8 41.40167 15.95785
Interpretation: The grand mean for factor A is 41.40 with CV of 15.95%
$parameters
test name.t ntr alpha
Duncan Fa 5 0.05
$duncan
Table CriticalRange
2 3.261182 5.563160
3 3.398460 5.797338
4 3.475191 5.928231
5 3.521194 6.006707
Interpretation: The LSD value is 5.563
$means
ch2 std r Min Max Q25 Q50 Q75
Bishnu 44.84667 6.017778 15 35.21 53.61 40.0600 45.21 50.310
Bramha 43.63000 7.290326 15 25.61 53.81 40.7100 43.11 48.910
Ganesh 36.33000 5.838910 15 27.71 48.91 31.5475 36.41 38.535
Laxmi 47.36000 5.680229 15 38.66 56.01 43.4100 46.81 51.835
Riddi 34.84167 7.739812 15 22.61 54.31 30.1100 34.21 38.860
$comparison
NULL
$groups
ch2 groups
Laxmi 47.36000 a
Bishnu 44.84667 a
Bramha 43.63000 a
Ganesh 36.33000 b
Riddi 34.84167 b
attr(,"class")
[1] "group"
Interpretation: The maximum chlorophyll content was seen for Laxmi variety which is significantly higher than Ganesh and Riddi but is at par with Bishnu and Bramha.
b. For Split plot (pesticide)
> out1<-with(split,duncan.test(ch2,Fb,DFerror = 40,MSerror = 30.20))
> out1
$statistics
MSerror Df Mean CV
30.2 40 41.40167 13.27351
$parameters
test name.t ntr alpha
Duncan Fb 5 0.05
$duncan
Table CriticalRange
2 2.858232 4.055602
3 3.005302 4.264283
4 3.101506 4.400788
5 3.170852 4.499184
$means
ch2 std r Min Max Q25 Q50 Q75
Azadirachtin 40.54000 8.020833 15 22.61 53.81 36.7725 42.31 46.06
Control 40.03833 7.064365 15 29.61 52.36 34.6600 38.66 44.71
Cow's milk 39.86333 8.798746 15 24.91 52.11 34.4850 40.01 45.11
Imidacloprid 47.59000 7.623760 15 30.61 56.01 41.9100 51.51 52.91
Jholmal 38.97667 6.552281 15 25.61 51.11 35.0100 40.61 42.76
$comparison
NULL
$groups
ch2 groups
Imidacloprid 47.59000 a
Azadirachtin 40.54000 b
Control 40.03833 b
Cow's milk 39.86333 b
Jholmal 38.97667 b
attr(,"class")
Interpretation: The chlorophyll content is significantly higher in Imidacloprid sprayed plot than all other treatments.
c. For interaction
> out2<-with(split,duncan.test(ch2,Fa*Fb,DFerror = 40,MSerror = 30.20))
> out2
Note: I have not shown it here as significance was not observed in this case.
OR
You need to first assign gla as model and gl.a is combined with dollar operator to represent error degree of freedom (8) for main plot factor. Similarly, assign glb to represent error degree of freedom (40) for the subplot factor and interaction term.
> gla<-aov$gl.a
> glb<-aov$gl.b
> Ea<-aov$Ea
> Eb<-aov$Eb
Now, run the command as shown below:
> out1<-with(split,duncan.test(ch2,Fa,gla,Ea,console=TRUE))
> out2<-with(split,duncan.test(ch2,Fb,glb,Eb,console=TRUE))
> out3<-with(split,duncan.test(ch2,Fa:Fb,glb,Eb,console=TRUE))
We can also see error and bar graphs as shown below;
> hist(ch2)
> bar.err(out1$means,variation="SE",ylim=c(0,60))

We can also see error and bar graphs as shown below;
> hist(ch2)
> bar.err(out1$means,variation="SE",ylim=c(0,60))
> bar.err(out2$means,variation="SE",ylim=c(0,60))
> bar.err(out3$means,variation="SE",ylim=c(0,60))
> bar.err(out3$means,variation="SE",ylim=c(0,60))
R commands for strip plots
File used: split.xls https://drive.google.com/file/d/1eGIeKlXWHpnPqeTiihsVcGIQAznM6jZu/view?usp=sharing
Note: Perform the first three commands as given for split plot.
1. Run ANOVA for strip plots
> aov<-with(split,strip.plot(Rep,Fa,Fb,ch2))
ANALYSIS STRIP PLOT: ch2
Class level information
Fa : Ganesh Bramha Riddi Bishnu Laxmi
Fb : Jholmal Azadirachtin Imidacloprid Cow's milk Control
Rep : R1 R2 R3
Number of observations: 75
model Y: ch2 ~ Rep + Fa + Ea + Fb + Eb + Fb:Fa + Ec
Analysis of Variance Table
Response: ch2
Df Sum Sq Mean Sq F value Pr(>F)
Rep 2 163.45 81.73 2.4980 0.098158 .
Fa 4 1816.36 454.09 10.4031 0.002944 **
Ea 8 349.20 43.65 1.3342 0.262717
Fb 4 737.16 184.29 9.1637 0.004412 **
Eb 8 160.89 20.11 0.6147 0.758865
Fb:Fa 16 561.12 35.07 1.0719 0.417744
Ec 32 1046.94 32.72
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
cv(a) = 16 %, cv(b) = 10.8 %, cv(c) = 13.8 %, Mean = 41.40167
The mean separation is done by DMRT (duncan.test). Other options can also be used. For main treatment factors, I have used error mean square and error degree of freedom respective to factor A and B from analysis of variance table. Similarly, for interaction error C for degree of freedom and its respective error mean square is used to compute.
The mean separation is done by DMRT (duncan.test). Other options can also be used. For main treatment factors, I have used error mean square and error degree of freedom respective to factor A and B from analysis of variance table. Similarly, for interaction error C for degree of freedom and its respective error mean square is used to compute.
For
this objective, I assigned gla as model and gl.a is combined
with dollar operator to represent error degree of freedom (8) for the
first factor. Similarly, assigned glb to represent error
degree of freedom (8) for the second factor. Then assigned glc to represent third
error degree of freedom (32) for the interaction term.
> gla<-model$gl.a
> glb<-model$gl.b
> glc<-model$gl.c
Moreover,
for error mean square Ea was assigned as model and was combined with dollar
operator. This gave the value of error mean square (23.734) for first factor.
Similarly, Eb was assigned for representing error mean square (17.806) for second
factor. Ec was assigned to represent error mean square (18.763) for
interaction.
> Ea<-model$Ea
> Eb<-model$Eb
> Ec<-model$Ec
2. Now the mean separation can be done as follows:
> out1<-with(split,duncan.test(ch2,Fa,gla,Ea))
> out1
If you don't want to do the steps mentioned above, you can do the following steps:
> out1<-with(split,duncan.test(ch2,Fa,gla,Ea))
> out1
If you don't want to do the steps mentioned above, you can do the following steps:
> out1<-with(split,duncan.test(ch2,Fa,aov$gl.a,aov$Ea))
> out1
$statistics
MSerror Df Mean CV
43.64948 8 41.40167 15.95775
$parameters
test name.t ntr alpha
Duncan Fa 5 0.05
$duncan
Table Critical Range
2 3.261182 5.563127
3 3.398460 5.797304
4 3.475191 5.928196
5 3.521194 6.006671
$means
ch2 std r Min Max Q25 Q50 Q75
Bishnu 44.84667 6.017778 15 35.21 53.61 40.0600 45.21 50.310
Bramha 43.63000 7.290326 15 25.61 53.81 40.7100 43.11 48.910
Ganesh 36.33000 5.838910 15 27.71 48.91 31.5475 36.41 38.535
Laxmi 47.36000 5.680229 15 38.66 56.01 43.4100 46.81 51.835
Riddi 34.84167 7.739812 15 22.61 54.31 30.1100 34.21 38.860
$comparison
NULL
$groups
ch2 groups
Laxmi 47.36000 a
Bishnu 44.84667 a
Bramha 43.63000 a
Ganesh 36.33000 b
Riddi 34.84167 b
attr(,"class")
[1] "group"
> out6<-with(split,duncan.test(ch2,Fb,aov$gl.b,aov$Eb))
> out6
$statistics
MSerror Df Mean CV
20.11077 8 41.40167 10.8317
$parameters
test name.t ntr alpha
Duncan Fb 5 0.05
$duncan
Table CriticalRange
2 3.261182 3.776103
3 3.398460 3.935056
4 3.475191 4.023902
5 3.521194 4.077169
$means
ch2 std r Min Max Q25 Q50 Q75
Azadirachtin 40.54000 8.020833 15 22.61 53.81 36.7725 42.31 46.06
Control 40.03833 7.064365 15 29.61 52.36 34.6600 38.66 44.71
Cow's milk 39.86333 8.798746 15 24.91 52.11 34.4850 40.01 45.11
Imidacloprid 47.59000 7.623760 15 30.61 56.01 41.9100 51.51 52.91
Jholmal 38.97667 6.552281 15 25.61 51.11 35.0100 40.61 42.76
$comparison
NULL
$groups
ch2 groups
Imidacloprid 47.59000 a
Azadirachtin 40.54000 b
Control 40.03833 b
Cow's milk 39.86333 b
Jholmal 38.97667 b
attr(,"class")
[1] "group"
>
> out7<-with(split,duncan.test(ch2,Fa:Fb,aov$gl.c,aov$Ec))
> out7
$statistics
MSerror Df Mean CV
32.71685 32 41.40167 13.81554
$parameters
test name.t ntr alpha
Duncan Fa:Fb 25 0.05
$duncan
Table CriticalRange
2 2.880659 9.512988
3 3.027689 9.998535
4 3.123222 10.314019
5 3.191582 10.539771
6 3.243335 10.710677
7 3.283984 10.844914
8 3.316744 10.953101
9 3.343648 11.041949
10 3.366058 11.115952
11 3.384925 11.178259
12 3.400942 11.231153
13 3.414625 11.276340
14 3.426369 11.315123
15 3.436482 11.348519
16 3.445208 11.377337
17 3.452745 11.402225
18 3.459253 11.423716
19 3.464864 11.442247
20 3.469690 11.458183
21 3.473823 11.471831
22 3.477342 11.483453
23 3.480344 11.493367
24 3.482802 11.501485
25 3.484829 11.508179
$means
ch2 std r Min Max Q25 Q50 Q75
Bishnu:Azadirachtin 44.01833 6.134958 3 37.135 48.910 41.5725 46.010 47.4600
Bishnu:Control 42.51833 4.729715 3 38.510 47.735 39.9100 41.310 44.5225
Bishnu:Cow's milk 49.01000 5.026927 3 43.210 52.110 47.4600 51.710 51.9100
Bishnu:Imidacloprid 50.24333 4.441096 3 45.210 53.610 48.5600 51.910 52.7600
Bishnu:Jholmal 38.44333 2.853653 3 35.210 40.610 37.3600 39.510 40.0600
Bramha:Azadirachtin 46.44333 6.395571 3 42.310 53.810 42.7600 43.210 48.5100
Bramha:Control 41.71000 5.436911 3 35.710 46.310 39.4100 43.110 44.7100
Bramha:Cow's milk 45.97667 6.051722 3 40.010 52.110 42.9100 45.810 48.9600
Bramha:Imidacloprid 48.07667 6.384617 3 40.710 52.010 46.1100 51.510 51.7600
Bramha:Jholmal 35.94333 8.957864 3 25.610 41.510 33.1600 40.710 41.1100
Ganesh:Azadirachtin 34.68500 4.469550 3 29.610 38.035 33.0100 36.410 37.2225
Ganesh:Control 33.39333 1.834621 3 31.460 35.110 32.5350 33.610 34.3600
Ganesh:Cow's milk 32.45167 5.229145 3 27.710 38.060 29.6475 31.585 34.8225
Ganesh:Imidacloprid 43.67667 4.974267 3 39.010 48.910 41.0600 43.110 46.0100
Ganesh:Jholmal 37.44333 6.390879 3 31.510 44.210 34.0600 36.610 40.4100
Laxmi:Azadirachtin 45.67667 2.676440 3 42.810 48.110 44.4600 46.110 47.1100
Laxmi:Control 47.49333 7.663115 3 38.660 52.360 45.0600 51.460 51.9100
Laxmi:Cow's milk 41.57667 2.478575 3 39.810 44.410 40.1600 40.510 42.4600
Laxmi:Imidacloprid 54.74333 2.193931 3 52.210 56.010 54.1100 56.010 56.0100
Laxmi:Jholmal 47.31000 3.576311 3 44.010 51.110 45.4100 46.810 48.9600
Riddi:Azadirachtin 31.87667 8.405554 3 22.610 39.010 28.3100 34.010 36.5100
Riddi:Control 35.07667 5.947549 3 29.610 41.410 31.9100 34.210 37.8100
Riddi:Cow's milk 30.30167 6.407239 3 24.910 37.385 26.7600 28.610 32.9975
Riddi:Imidacloprid 41.21000 12.046161 3 30.610 54.310 34.6600 38.710 46.5100
Riddi:Jholmal 35.74333 4.572016 3 31.710 40.710 33.2600 34.810 37.7600
$comparison
NULL
$groups
ch2 groups
Laxmi:Imidacloprid 54.74333 a
Bishnu:Imidacloprid 50.24333 ab
Bishnu:Cow's milk 49.01000 abc
Bramha:Imidacloprid 48.07667 abcd
Laxmi:Control 47.49333 abcd
Laxmi:Jholmal 47.31000 abcd
Bramha:Azadirachtin 46.44333 abcde
Bramha:Cow's milk 45.97667 abcdef
Laxmi:Azadirachtin 45.67667 abcdef
Bishnu:Azadirachtin 44.01833 abcdefg
Ganesh:Imidacloprid 43.67667 abcdefgh
Bishnu:Control 42.51833 bcdefghi
Bramha:Control 41.71000 bcdefghi
Laxmi:Cow's milk 41.57667 bcdefghi
Riddi:Imidacloprid 41.21000 bcdefghij
Bishnu:Jholmal 38.44333 cdefghij
Ganesh:Jholmal 37.44333 defghij
Bramha:Jholmal 35.94333 efghij
Riddi:Jholmal 35.74333 efghij
Riddi:Control 35.07667 fghij
Ganesh:Azadirachtin 34.68500 fghij
Ganesh:Control 33.39333 ghij
Ganesh:Cow's milk 32.45167 hij
Riddi:Azadirachtin 31.87667 ij
Riddi:Cow's milk 30.30167 j
attr(,"class")
[1] "group"
Interpretation: Try to do it yourself. Same letter signify that they are not significantly different.









1 Comments:
Thank you for this tutorial, it was a blessing
Post a Comment
Subscribe to Post Comments [Atom]
<< Home